Spotlight: Annette Govindarajan November 19, 2020
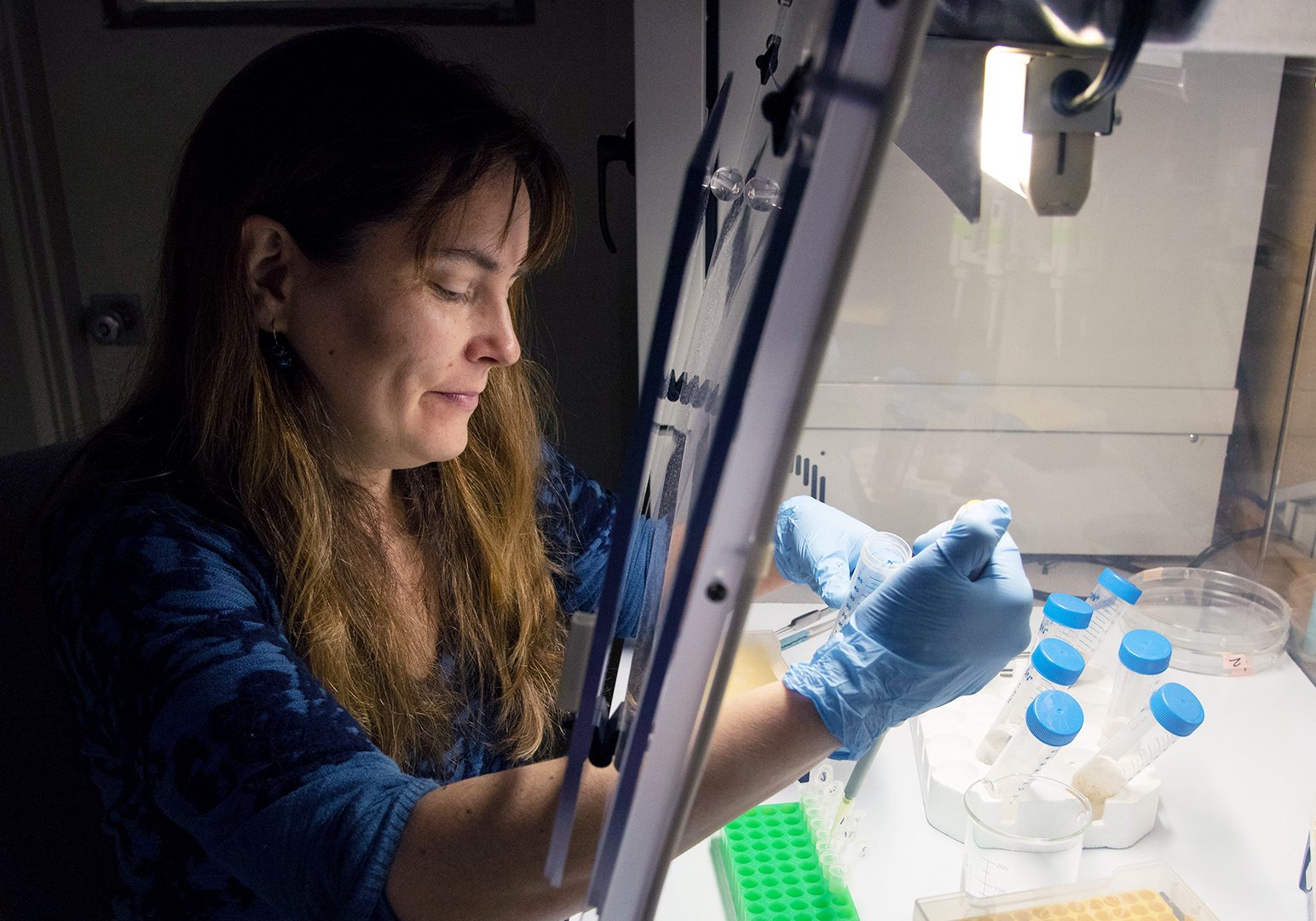
Annette Govindarajan studies the biodiversity, ecology and evolution of marine animals. As a principal investigator for the OTZ project, she examines genetic material in seawater (also called environmental DNA or “eDNA”) to learn more about organisms that are present in the deep ocean. It’s cutting-edge work, and it’s revealing some species that would be nearly impossible to find using other methods. In this interview, learn about Govindarajan’s work with eDNA, what keeps her passionate about this new technology, and more.
What kinds of questions are you trying to answer about the ocean twilight zone?
I'm investigating the kinds of animal biodiversity that are in the OTZ. For example, what species are there? Where do we find these species? What are the factors controlling their distribution? Those are some of the big unanswered questions. I think one of the most exciting aspects of this work is that it has the potential to help us discover new species in the OTZ that we didn’t realize were there
How does studying eDNA help us better understand the twilight zone ecosystem?
The beauty of eDNA is that you don’t need to take genetic samples from specimens - only seawater. If we find an animal’s DNA is in the water, we expect that it was present somewhere, even if we never actually see it. And that’s great, because some of the species that live in the OTZ are particularly hard to catch in a net and study directly, especially jellyfish and other gelatinous animals. They're so delicate that they fall apart if you try to collect them in a net. We actually think that gelatinous animals are underestimated in the OTZ—there are probably a lot more of them than our net sampling shows. By using eDNA, though, we hope to get a more comprehensive picture of what's down there. It’s a new tool in our toolkit, and it nicely compliments the other methods we’re using.
What are some of the main challenges of studying eDNA?
Given that it’s such a new technique for the mesopelagic, we still have a lot to learn about the best ways to use it. For example, more work needs to be done to understand how eDNA signals relate to actual animal abundances and biomass. Also, in order to put species names on the eDNA sequences that we find, we first have to match our sequences to a pre-existing genetic reference database. That means we need to flesh out the database in the first place with sequences from animals that we’ve already identified. Doing that can be pretty straightforward if you already have a physical specimen of animal to work with—but it can be a real challenge to get a positive ID for animals that are hard to catch, like gelatinous species.
You’re also working on ways to automate the sampling process. Why is that important?
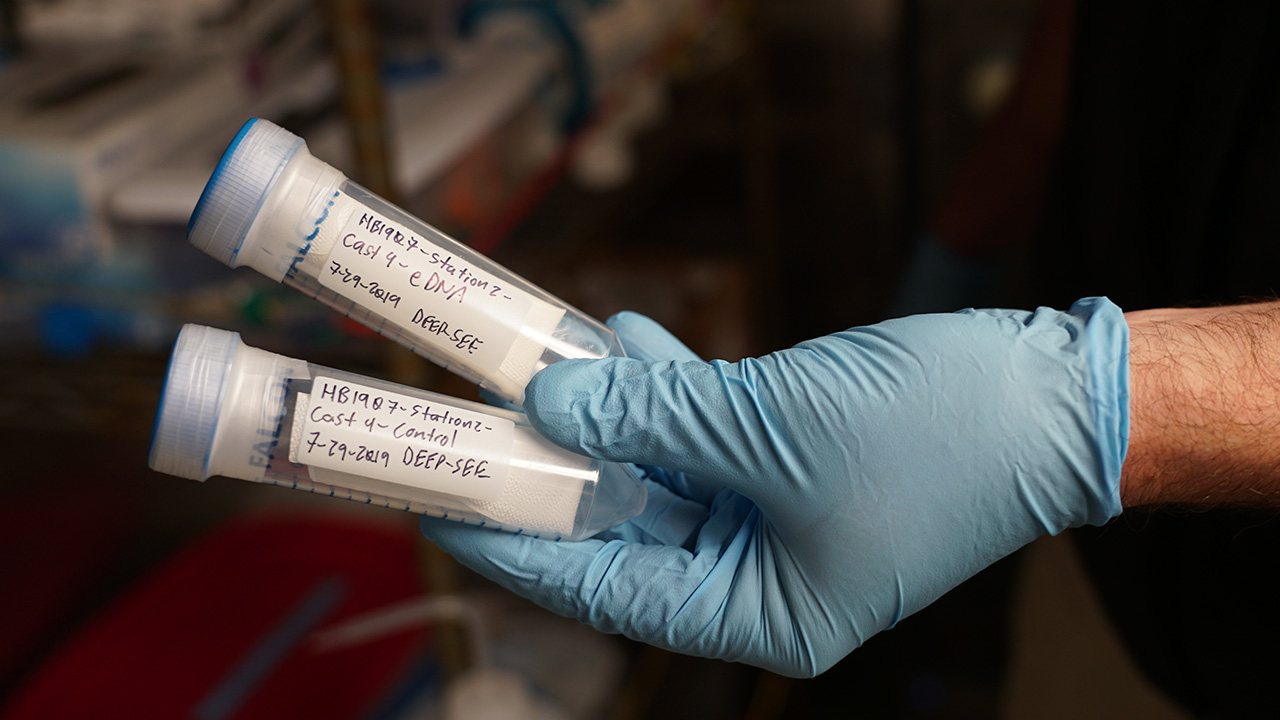
Getting good eDNA samples isn’t very easy. Typically, they’re collected by filling bottles (called Niskin bottles) in the deep ocean, pulling them up to the ship, and then filtering the water inside them to obtain the genetic material. It’s a long process, and when you have a dozen or more bottles, it can take a few hours. You also have to be really careful about contamination while you’re filtering, especially on a ship—there are usually lots of other scientists aboard who are working with their own samples, so we have to try to carve out our own space and sterilize it before each use. To get around that problem, we’re working on creating autonomous samplers that can filter seawater at depth, so when the sampler comes back up, we can just remove the filter and put it in the freezer for storage. Currently we’ve deployed our sampler with Mesobot, a new hybrid vehicle for studying the midwater. This approach gives us more flexibility in sampling, reduces the chances for contamination, and saves labor.

What is one of the most exciting things you’ve found in the OTZ so far?
On one cruise, we pulled up a bizarre looking blob in one of our large net tows. When I sequenced its DNA, I discovered it was a fragment of an extremely rare deep sea jellyfish—one that is only known to science thanks to a handful of records. So using genetic analysis, I was able to learn something new about the distribution of this species, and record a new sighting of it in a place where it hadn't been seen before. That's the kind of thing that I ultimately want to do with eDNA in conjunction with other approaches.
Why do you think it’s so important to study the OTZ now? What’s at stake?
Many scientists have studied coastal areas as well as the surface ocean and deep sea floor. But in between the surface and the bottom, there’s the vast middle part of the ocean, which is mind-bogglingly large. That's where the OTZ is located, and it’s an area of the ocean that we know very little about.
Over the last few years, some studies of the OTZ have suggested that there’s much more life down there than we once thought—as in an order of magnitude more. Much of that life could be harvestable using deep-sea fishing nets. There is a lot of interest in capturing deep-sea species on a commercial level, particularly as coastal species have declined. The problem is that we don’t yet have enough basic information about the OTZ to ensure that the species living there are harvested in a sustainable way. We’d need to know things like: what exact species are there? How long do they live? What's their life cycle like? How many offspring do they produce? Really, just basic questions about life history and biodiversity, We absolutely need to get some answers before people begin exploiting the OTZ—not after the fact when it’s too late.