eDNA Explained
What is eDNA?
Environmental DNA (eDNA) is essentially DNA collected from the environment.
As animals swim through the ocean, they're constantly releasing DNA as they shed skin or scales into the water column. DNA is also in sperm and eggs released during spawning, in the chunks of tissues that remain in the water column after a messy feeding session, and even in their fecal matter. In ocean environments, eDNA is potentially dispersed passively with currents, on sinking particles, and through biological facilitation (e.g., in the guts of other animals). OTZ researchers can then collect this eDNA from seawater samples, rather than from animals directly, letting them tell which species are present in the twilight zone—even if they never directly spot those animals.
What can eDNA tell us about biodiversity?
eDNA lets scientists detect animals present in an area without actually collecting the animals themselves. This can be a very efficient way to obtain a snapshot of the types of species that are present in a region, especially since many species are rare or difficult to collect using traditional methods like nets. Jellyfish, for example, are extremely delicate, and often fall apart in sampling nets. Likewise, many fish can sense the presence of nets as they move through the water, and can avoid being caught in the first place. eDNA analysis offers a valuable new tool to detect those species, providing a more nuanced understanding of biological communities.
How is eDNA studied?
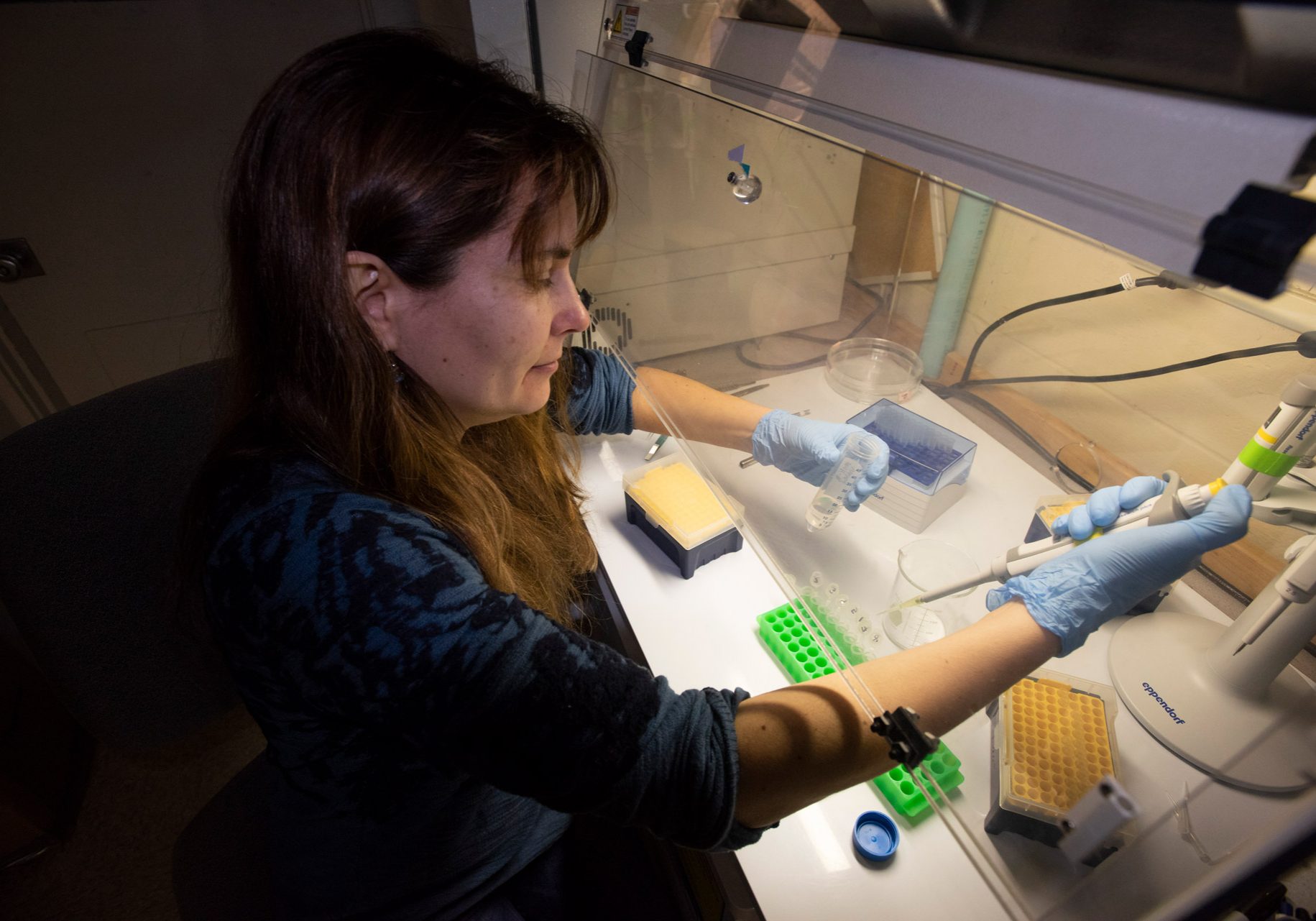
Scientists collect eDNA by filtering water samples through a sub-micron filter and extracting the DNA from the filter. The extracted DNA is usually processed in one of two ways. In the first way, genetic markers (or “DNA barcodes”) from different DNA traces in the sample are sequenced, and those sequences are then compared to a reference database. This helps to identify the species of animals present in the sample. Some genetic sequences are found in a broad range of species, while others indicate more specific groups, such as fishes. This approach is called “amplicon sequencing” or “metabarcoding”. The metabarcoding method can provide information about marine communities and and other co-existing species, or can be used to monitor how communities change in the face of human-caused stressors like climate change and increasing water temperatures.
Genetic markers from specific species can also be amplified, meaning many copies of that DNA fragment are generated and analyzed. This approach is called “qPCR” or “quantitative PCR” and is used for estimating how many of a particular species are present in the environment. The approach is useful for obtaining information about a specific endangered species, invasive species, or a species of economic value.
How are scientists using eDNA to learn more about biodiversity in the OTZ?
For the past three years, the OTZ team has been collecting and analyzing eDNA samples along vertical transects in the North Atlantic. These transects extend to the base of the mesopelagic zone (at roughly 1000 meters below the surface). The team is taking samples during the daytime and nighttime in order to detect diel vertical migration, when mesopelagic species migrate to shallow waters during the night and deeper waters during the day. Sampling is guided by shipboard acoustic sensors, which can indicate the presence of different numbers of organisms. The samples are sequenced using genetic metabarcoding approaches to detect a variety of animal types, including fish, crustaceans, and jellyfish.
Back in the laboratory, the team is also conducting experiments to understand eDNA shedding and decay, using a variety of diverse animal forms, temperatures, and light conditions that reflect the mesopelagic environment. They are also working to understand how eDNA moves throughout the twilight zone. OTZ scientists are currently developing new ways to collect eDNA autonomously using platforms like Mesobot (a robot designed for mesopelagic exploration) and Deep-See (a towed broadband acoustics and imaging instrument).
What are some of the challenges of studying eDNA?
Animal eDNA is present in very tiny quantities in the ocean, making it similar to finding a “needle in a haystack”. Because it occurs in such trace amounts, samples are easily contaminated during the collection process. Great care must be taken when gathering and processing samples, including includes sterilizing the work area and equipment, and running control samples to monitor for contamination. Trying to do this aboard a busy research ship with multiple other scientists working nearby is no small feat. Since eDNA analysis is a new approach, scientists are also still learning about a variety of factors that can affect their interpretations of eDNA signals, like the rates at which animals release eDNA, how quickly that eDNA degrades, and how eDNA is transported through the environment.